Image Segementation
Fully Convolutional Networks(FCN) for Semantic Segmentation
Main Takeaway
In this post, I will implement Fully Convolutional Networks(FCN) for semantic segmentation on MIT Scence Parsing data.
While object detection methods like R-CNN heavily hinge on sliding windows (except for YOLO),
FCN doesn’t require it and applied smart way of pixel-wise classification. To make it possible, FCN implements 3 distinctive features in their network.
Key Features
- Spatial Map
- Get Spatial map (heatmap) instead of non-spatial outpues by chaning Fully Connected Layer into 1 X 1 Convolution
- Deconvolution
- In-network Upsampling for pixel-wise prediction
- Skip Architecture
- Combine layers of the feature hierarchy to get local predictions with respect to global strucutre, also called as deep-jet
Details of Features
- Spatial Map With regard to “1. Spatial Map”, it allows us to get heatmap not vector output, which convert classification problem into coarse spatial map.
-
Deconvolution Upsampling the coarse spatial map into pixel of original image is performed through in-network nonlinear upsampling (DeConvolution). Since DeConvolutioon is implemented using reverse the forward and backward of convolution, the filter(e.g. ‘W_t1’) of DeConvoltion layer(‘tf.nn.conv2d_transpose’ in Tensorflow) can also be learned.
-
Skip Architecture
Finally, upsampling 32X generates srikingly coarse ouptuts. To overcome such phenomena, the author added the 2X upsample of last layer and the right before layer. This combination of layers of feature hierarchy expands the capabililty of network to make fine local prediction with respcet to global structures. The “fuse” in the code of figure corresponds to the combination of layers, and the combination procedure was performed up to 2 times that make us 8X upsamlple. Therefore, FCN-32s is converted to FCN-8s via Skip Architecture.
ReCap
- Modification
To train and validate the FCN-8s in Windows, I modified two scripts: read_MITSceneParsingData.py & FCN.py. First and foremost, the script read_MITSceneParsingData.py evokes an issue of parsing, which disables getting training and validation data. Last but not least, I changed the batch size from 2 to 10, which significantly reduced cost and ‘main’ function for validation (NVIDIA GPU Geforce 1080ti 11G is used).
- Results
One of random validation samples is listed below. The quality of prediction result is not precise as much as I expected. One possible reason is the number of classes in read_MITSceneParsing data (151), while the paper used PASCAL VOC 2011 data (21).
Reulsts in Paper
Reference
Resources that I referenced are listed at the bottom of this post.
- FCN semantic segmentation paper: FCN Paper
- Github code : FCN github
Bio-Chemical Literature Review
New Chemical Design/Property Prediction/Target DeConvoltion
List of papers
| category | Paper | Model | Takeaway |
|---|---|---|---|
| New Chemical Design | BenovolentAI | RNN based RL (HC-MLE) | Reinfocement Learning with 19 benchmark |
| VAE_property | VAE(1D CNN & RNN) wth property(GP) | Variatioanl AutoEncoder jointly predicting property | |
| ChemTS | MonteCarlo Tree Search with RNN | Cascading way and RNN is used at RollOut step | |
| DruGAN | Adversarial AutoEncoder | AAE is better than VAE in terms of Reconstruction error and Diversity | |
| InSilico | RNN-LSTM | Unique Scaffolds can be achieved | |
| Property Prediction | DeepChem | Graph Convolution | Considering 2D strucutre is critical |
| Target DeConvoltion | SwissTarget | Logist Regression | 3D similarity based on ligands |
To be compact, I didn’t upload Input, related property, and data resources though being created. If somebody pursue for more details about each paper, please feel free to email me.
Algorithms
For the sake of compactness, I would demonstrate two methods Hillclimb MLE (HC-MLE) in Reinfocement Learning and Monte Carlo Tree Search from BenovolentAI and ChemTS papers. The second paper, VAE with Property, is reviewed in my previous post.
1. Hillclimb MLE (HC-MLE)
First, There are 19 benchmarks that used for Reward in Reinforcement Learning. They can be catagorized into Validity, Diversity, Physio-Chemical Property, similarity with 10 representative compounds, Rule of 5, and MPO. Secondly, HC-MLE mazimizes the likelihood of sequences that received Top K highest reward.
2. Monte Carlo Tree Search (MCTS)
- Pretrain RNN and Get Conditional Probability
- Conditional Probability is used in MCTS as sampling distribution to get next character and elongate the smiles code
- Reward score of generated smiles code is computed
- In Back propagation, reward is back propagated & UCB at each node is updated
Reference
- benevolent (EXPLORING DEEP RECURRENT MODELS WITH REINFORCEMENT LEARNING FOR MOLECULE DESIGN): benevolent
- VAE with Property (Automatic Chemical Design Using a Data-Driven Continuous Representation of Molecules): vae_property
- ChemTS (Python Library): ChemTS
- druGAN (druGAN: An Advanced Generative Adversarial Autoencoder Model 2 for de Novo Generation of New Molecules with Desired Molecular 3 Properties in Silico ): druGAN
- InSilico (In silico generation of novel, drug-like chemical matter using the LSTM neural network ): InSilico
-
DeepChem (Applying Automated Machine Learning to Drug Discovery) : N/A
-
SwissTarget (SwissTargetPrediction: a web server for target prediction of bioactive small molecules ): SwissTarget
De Novo Design: Derivatives of Autoencoder
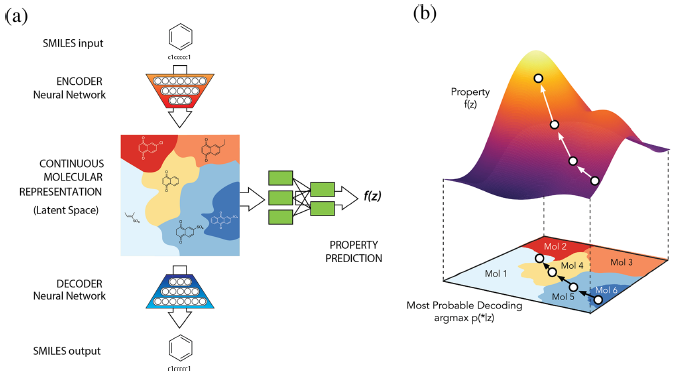
A primary goal of designing a deep learning architecture is to restrict the set of functions that can be learned to ones that match the **desired properties** from the domain
S Kearnes etal 2016 (Molecular Grpah Convolutions: Moving Beyond Fingerprints)
When building deep learning architecture, the above statement strongly highlights the key concept of implementing deep learning architecture. Especially, phrase “match the desired properties” can be understood as teaching, imposing a constraint, or regularization in several learning algorithm. For instance, GAN attempts to impose regularization by introducing Discriminator & Generator on the purpose of setting, and derivatives of Autoencoder predict a certain property by attaching MLP from latent space to get new datum with desired properties. Following takeaway outlines what I will illustrate in series with respect to generative models.
Structure of Generative Models
As Ian Goodfellow (NIPS 2014, and 2016 tutorial) demonstrates that maximizing Maximum Likelihood Estimates is equivalent to minimizing KL divergence between data generating distribution and the model and introduces Nash equilibrium, which verifies the unbiasness of GAN. On top of that, Generating a datum via Variational Autoencoder while simultaneously predicing property can be regarded as optimization with constraint. whether explicitily represent a probability distribution or not derives the fundamental difference between these two generative models(GAN & Variational Autoencoder).
Although the way of approach is different, convergence of disciplines such as Economcis, Industrial Engineering, Statistics, and Computer Science has appeared and they share the goal of implementing natural “regularization”.
As for the widley known argloithm, foramts of regularization can be categorized into 2 folds: Game(Adversarial) and Predict property simultaneously(Constraint). By starting from the perspective of Autoencoder, I made 3 overlapping diagrams where “Adversarial” and “Constraint” lie on the domain of “regularization”.
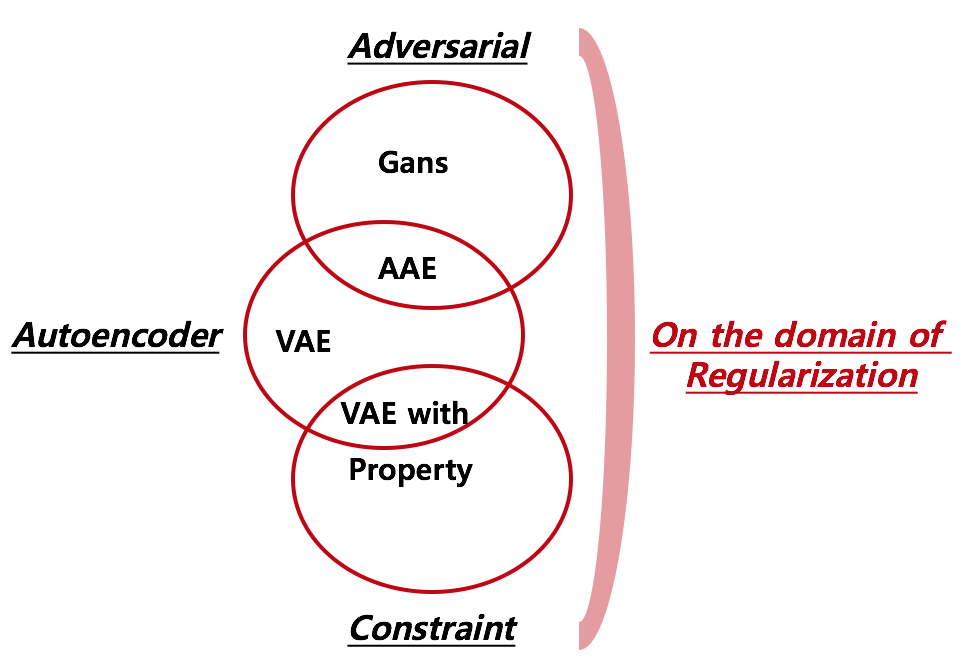
1. Comparison of Models
Below picture is combination of figures from different papers. Red box indicated in derivatives of autoencoder denotes different substructure across derivatives.
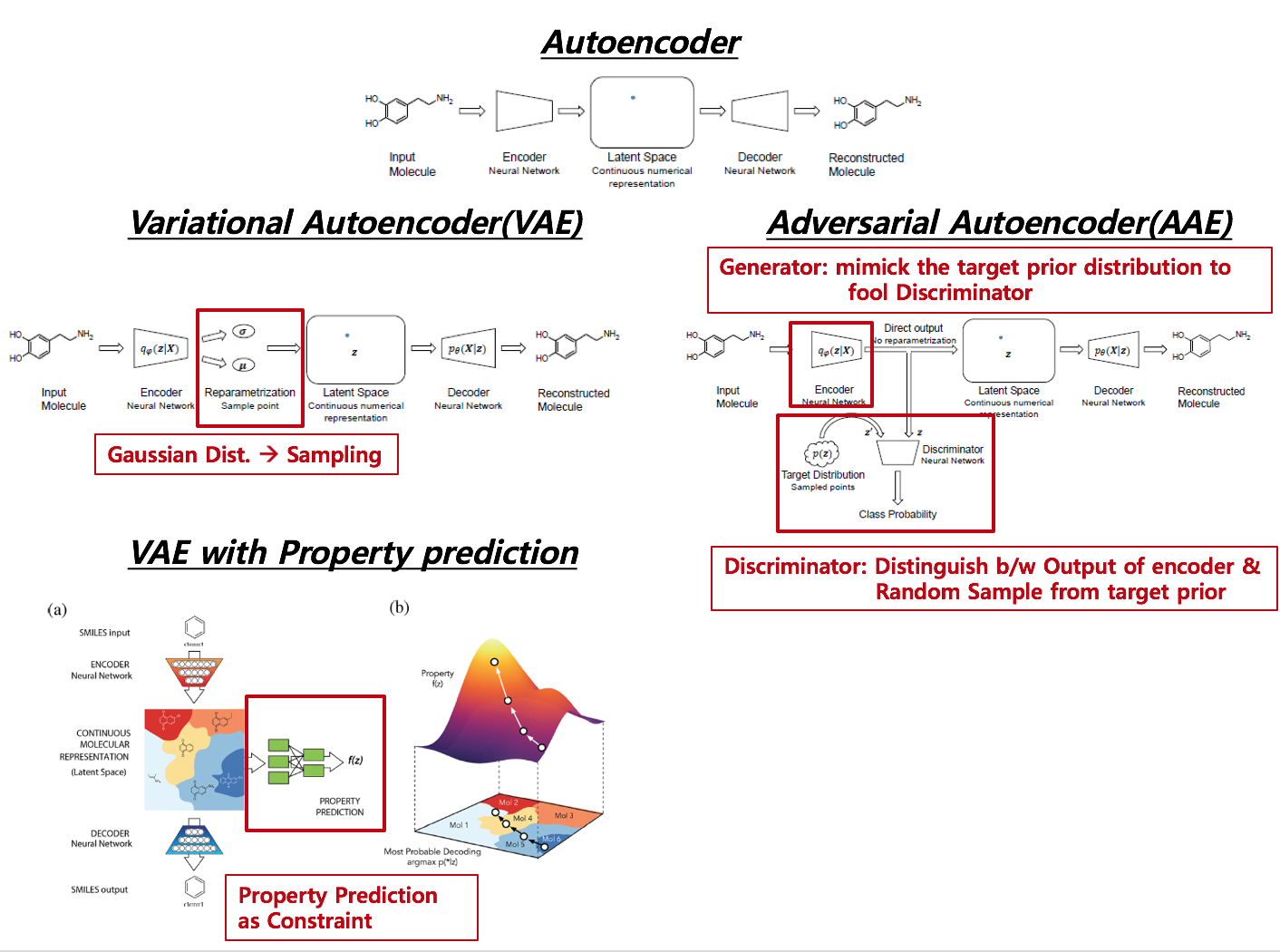
Distinctive Features across Methods
-
AE –> VAE Reparametrization from encoder to Latent Space via pre-assumed distribution (usually Gaussian distribtion)
-
VAE –> VAE with Property Added a netwrok from latent space to a supervised network such as Regression or Classification, which agglomerates Latent Space according to the level of target value.
-
AE –> AAE No Reparametrization, but adversarial Network is attached
Cost function across Methods
- AE : Reconstruction error
- VAE : Reconstruction error + KL-divergence (Regularizing Variational Parameter)
( latent space is driven by adding stochasticity to the encoder ) –> Adding noise to the encoded latent space forces the decoder to learn a wider range of latent points, which lead to find out more robust representations. - VAE with property: Reconstruction error of decoder + KL-divergence (Regularizing Variational Parameter) + regression error
- AAE : Reconstruction error of decoder + Discriminator loss + encoder(Generator) loss
In terms of regularizing the encoder posterior to match a pre-assumed prior, VAE with property and Adversarial autoencoder are on the same domain.
Specifically, Discriminator and Generator loss can be implemented like following
Discriminator related loss:
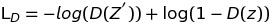
Generator related loss:
2. Data & Pre-processing
- Image or sequence (Molecular information is denoted as SMILES code, which is a sequence, will be mainly used )
In this post, I will use data from Tox21 data and the input will be SMILES code, which is mainly used for chemical design and molecular property prediction. Data pre-processing and specific model illustration is described below ( reference: T Blaschke etal 2017 ).
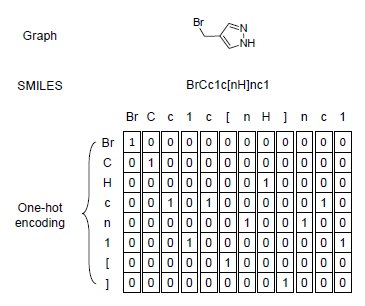
To adopt periodic table into SMILES code, I merged chemical in periodic table with the code and generated one-hot encoded matrix for each SMILES code. Since the max length of chemical is 2, ismol variable in the function is
# tscode : SMILES code
# chnum : Total unique number of chemcial in SMILES code considering periodic table
# ohlen : Maximum number of SMILES code length (Usually restricted by researchers and users)
def onehot_encoder(tscode, chnum, ohlen):
ohcode = np.zeros( ( chnum * ohlen ) )
ohindex = 0
mm = 0
while mm < len( tscode ) :
ch0 = tscode[ mm ]
ismol = 0
if ch0 in mcode :
ismol = 1
if mm < len( tscode ) - 1 :
ch1 = tscode[ mm + 1 ]
if ch0 + ch1 in mcode :
ismol = 2
mm = mm + 1
if ismol == 0 :
ch = ch0
elif ismol == 1 :
ch = ch0
elif ismol == 2 :
ch = ch0 + ch1
indx = codelist.index( ch )
ohcode[ ohindex + indx ] = 1
mm = mm + 1
ohindex = ohindex + chnum
return ohcode3. Implementation of VAE with Proprty
3.1 HyperParameter Setting
import numpy as np
import pandas as pd
import tensorflow as tf
# Convert smiles code into 2D array
import smiles_onehot_func as sof
from math import ceil
from datetime import datetime
from rdkit.Chem import MolFromSmiles
from rdkit.Chem.QED import qed
from rdkit.Chem.Descriptors import MolWt
from rdkit import DataStructs
from rdkit.Chem.Fingerprints import FingerprintMols
scode = sof.trX
ver_str = "v1.1.3"
# GPU allocation
gpu_num = str( 0 )
learning_rate = 0.0001
num_epoch = 1000
num_rs = 5000
batch_size = 50
print_time = 10
save_epoch = 10
# Latent Space dimension in AE
latent_dim = 1024
# Latent Space dimension in VAE (latent dim --> variational_dim via Reparametrization trick)
variation_dim = 256
# 2D array dimension
ohlen = sof.m_ohlen
chnum = sof.m_chnum
# Convolution nodes
num_clayer1 = 64
num_clayer2 = 128
ol_devide_2 = ceil( ohlen / 2 )
ol_devide_4 = ceil( ol_devide_2 / 2 )
cl_devide_2 = ceil( chnum / 2 )
cl_devide_4 = ceil( cl_devide_2 / 2 )
# Placeholders for Input and target
x = tf.placeholder( tf.float32, [ None, ohlen, chnum, 1 ], name="input" )
label = tf.placeholder( tf.float32, [ None, ohlen, chnum, 1 ], name="label" )
latent = tf.placeholder( tf.float32, [ None, variation_dim ], name="latent" ) #latent space at VAE
prop = tf.placeholder( tf.float32, [ None, 1 ], name="prop" ) # regression target value
# Variational Parameters
weights = {
'z_mean': tf.Variable(tf.truncated_normal([latent_dim, variation_dim])),
'z_std': tf.Variable(tf.truncated_normal([latent_dim, variation_dim]))
}
biases = {
'z_mean': tf.Variable(tf.zeros([variation_dim])),
'z_std': tf.Variable(tf.zeros([variation_dim]))
}3.2 Encoder
def encoder( inputs, is_training ) :
# === Convoltion Sets
conv1 = tf.layers.conv2d( inputs = inputs, filters = num_clayer1, kernel_size = ( 3, 3 ), padding = 'same', activation = None )
conv1 = tf.layers.batch_normalization( conv1, center = True, scale = True, training = is_training )
conv1 = tf.nn.relu( conv1 )
maxpool1 = tf.layers.max_pooling2d( conv1, pool_size = ( 2, 2 ), strides = ( 2, 2 ) , padding = 'same' ) # 50 x 71
conv2 = tf.layers.conv2d( inputs = maxpool1, filters = num_clayer2, kernel_size = ( 3, 3 ), padding = 'same', activation = None )
conv2 = tf.layers.batch_normalization( conv2, center = True, scale = True, training = is_training )
conv2 = tf.nn.relu( conv2 )
maxpool2 = tf.layers.max_pooling2d( conv2, pool_size = ( 2, 2 ), strides = ( 2, 2 ) , padding = 'same' ) # 25 x 36
maxpool2 = tf.reshape( maxpool2, [ - 1, ol_devide_4 * cl_devide_4 * num_clayer2 ] )
encoded = tf.layers.dense( inputs = maxpool2, units = latent_dim, activation = None )
encoded = tf.layers.batch_normalization( encoded, center = True, scale = True, training = is_training )
encoded = tf.nn.relu( encoded, name = "encoded" )
# === ADD VARIATIONAL
z_mean = tf.matmul(encoded, weights['z_mean']) + biases['z_mean']
z_std = tf.matmul(encoded, weights['z_std']) + biases['z_std']
z_std = 1e-6 + tf.nn.softplus( z_std )
# === Reparametrization
z = z_mean + z_std * tf.random_normal(tf.shape(z_mean), 0, 1, dtype=tf.float32)
return z , z_mean, z_std3.3 Decoder
def decoder( encoded, is_training ) :
# === DeConvoltion Sets (More or less Upsampling)
fullcon = tf.layers.dense( inputs = encoded, units = ol_devide_4 * cl_devide_4 * num_clayer2, activation = None )
fullcon = tf.layers.batch_normalization( fullcon, center = True, scale = True, training = is_training )
fullcon = tf.nn.relu( fullcon )
fullcon = tf.reshape( fullcon, [ - 1, ol_devide_4, cl_devide_4, num_clayer2 ] )
upsample1 = tf.image.resize_images( fullcon, size = ( ol_devide_2, cl_devide_2 ), method = tf.image.ResizeMethod.NEAREST_NEIGHBOR )
conv4 = tf.layers.conv2d( inputs = upsample1, filters = num_clayer2, kernel_size = ( 3, 3 ), padding = 'same', activation = None )
conv4 = tf.layers.batch_normalization( conv4, center = True, scale = True, training = is_training )
conv4 = tf.nn.relu( conv4 )
upsample2 = tf.image.resize_images( conv4, size = ( ohlen, chnum ), method = tf.image.ResizeMethod.NEAREST_NEIGHBOR )
conv5 = tf.layers.conv2d( inputs = upsample2, filters = num_clayer1, kernel_size = ( 3, 3 ), padding = 'same', activation = None )
conv5 = tf.layers.batch_normalization( conv5, center = True, scale = True, training = is_training )
conv5 = tf.nn.relu( conv5 )
logits = tf.layers.conv2d( inputs = conv5, filters = 1, kernel_size = ( 3, 3 ), padding = 'same', activation = None )
logits = tf.add( logits, 0.0, name = "decoded" )
return logits3.4 Regression
def property_regression( encoded ) :
fullcon1 = tf.layers.dense( inputs = encoded , units = 512, activation = None )
fullcon1 = tf.nn.relu( fullcon1 )
fullcon2 = tf.layers.dense( inputs = fullcon1, units = 512, activation = None )
fullcon2 = tf.nn.relu( fullcon2 )
output = tf.layers.dense( inputs = fullcon2, units = 1 , activation = tf.nn.sigmoid )
output = tf.add( output, 0, name = "property" )
return output3.5 VAE Loss
def vae_loss(x_reconstructed, x_true, z_mean, z_std ):
# === Reconstruction loss
marginal_likelihood = tf.reduce_sum(tf.nn.sigmoid_cross_entropy_with_logits( logits=x_reconstructed, labels=x_true), axis=1)
# === VAE loss from another script file
KL_divergence = 0.5 * tf.reduce_sum(tf.square(z_mean) + tf.square(z_std) - tf.log(1e-8 + tf.square(z_std)) - 1, 1)
marginal_likelihood = tf.reduce_mean(marginal_likelihood)
KL_divergence = tf.reduce_mean(KL_divergence)
loss = marginal_likelihood + KL_divergence
return loss, marginal_likelihood, KL_divergence3.6 Training
def train( re_training = False ) :
# === Collect Variables via scope
with tf.variable_scope("AE"):
z, z_mean, z_std = encoder( x , is_training = True )
logits = decoder( z, is_training = True )
with tf.variable_scope( "FC" ) :
prop_reg = property_regression( z )
# === VAE loss
loss_ae , NML, KL = vae_loss( logits, label, z_mean, z_std )
# === Regression loss
loss_fc = tf.reduce_mean( tf.losses.mean_squared_error( labels = prop, predictions = prop_reg ) )
# === get variable for each model
t_vars = tf.trainable_variables( ) # return list
en_vars = [ var for var in t_vars if "AE" in var.name ]
# === Cost function
cost_ae = tf.add( loss_ae, 0, name = "cost_ae" )
cost_all = tf.add( cost_ae, loss_fc, name = "cost_all" )
# === Recall Batch Normalization in VAE
update_ops_en = tf.get_collection( tf.GraphKeys.UPDATE_OPS, scope = "AE" )
# === Optimization
optimizer = tf.train.AdamOptimizer( learning_rate )
with tf.control_dependencies( update_ops_en ):
optimizer_ae = optimizer.minimize( cost_all, var_list = en_vars, name = "optimizer_ae" )
init = tf.global_variables_initializer( )
saver = tf.train.Saver( )
c = tf.ConfigProto()
c.gpu_options.visible_device_list = gpu_num
with tf.Session( config = c ) as sess :
sess.run( init )
if re_training :
saver.restore( sess, save_path = tf.train.latest_checkpoint( model_path ) )
last_epoch = tf.train.latest_checkpoint( model_path ).split( "-" )[ - 1 ]
for epoch in range( num_epoch ) :
if re_training :
if epoch < int( last_epoch ) :
print( "skip epoch {}...".format( epoch ) )
continue
for i in range( num_rs ) :
batch, props = sof.GetOnehotBatch( batch_size )
imgs = batch.reshape( [ - 1, ohlen, chnum, 1 ] )
cs_ae, cs_nml, cs_kl, cs_fc, _ = sess.run( [ cost_ae, NML, KL ,loss_fc, optimizer_ae ],
feed_dict={ x : imgs, label : imgs, prop : props } )
if i % print_time == 0 :
print( ver_str + " Max Length : " + str( ohlen ) + " Using GPU : " + gpu_num )
print( "Epoch : {}/{} iter : {}/{}...".format( epoch + 1, num_epoch, i + 1, num_rs ) + "\n" +
"VAE loss : {:.6f} ".format( cs_ae ) +
"NML loss : {:.6f} ".format( cs_nml ) +
"KL loss : {:.6f} ".format( cs_kl ) +
" Prop Regression loss : {:.6f}".format( cs_fc ) +
" duration time : " + str( t4 - t3 )
if ( epoch + 1 ) % save_epoch == 0 :
saver.save( sess, model_path, global_step = ( epoch + 1 ) )
saver.save( sess, model_path, global_step = ( epoch + 1 ) )Reference
-
Kearnes, Steven, et al. “Molecular graph convolutions: moving beyond fingerprints.” Journal of computer-aided molecular design 30.8 (2016): 595-608.
-
Blaschke, Thomas, et al. “Application of generative autoencoder in de novo molecular design.” Molecular informatics 37.1-2 (2018).
-
Gómez-Bombarelli, Rafael, et al. “Automatic chemical design using a data-driven continuous representation of molecules.” ACS Central Science 4.2 (2018): 268-276.
-
Goodfellow, Ian, et al. “Generative adversarial nets.” Advances in neural information processing systems. 2014.
-
Github of VAE with property prediction : Chemical VAE
Deep Learning with Database as Executable file
Deep Learning Model + SQL Database + Threading
The task that I encountered was to construct a program, which can run by itself and should keep watching multiple Database tables at different time intervals. To make it possible, I used the thread with TCP/IP to check whether new data tables are updated in SQL. If new data is found, then I access the data while remembering previously processed one. After transforming the newly scanned data into a 2D matrix and projecting it as an image, I implemented trained CNN model to predict the grade of a product. Finally, I write the prediction result with corresponding information on an outcome table and saved the image into the path(written in JSON file).
Structure of Codes
1. Prerequisites
- Global Variables: Required information to access SQL
- JSON file: Paths where to save image and where trained model parameter located
2. Classes
- Class A: SQL manager
- Class B: Deep Learning Model and Pre-Processing
- Function C: scheduler, which calls the functions in Class A and Class B to check whether a new data came in and execute to save the image and perform prediction.
3. Main function
- Class A initialization enables us to connect database.
- Class B initialization read JSON file for necessary paths
- SetModel function in Class B ensures whether trained model parameters are loaded
- Catch for Function C
import treading
import logging
# 1.1 Global Varaibles such as localhost, id, and password to access SQL database
sql_host = 'tcp:xxx,xxx,xx,xx'
sql_user = 'wonik'
sql_password = 'jang'
sql_db = 'pro'
sql_encode = 'utf-8'
# 1.2 Load Json file, which contains relevant paths
abs_path = './'
json0 = 'path_inform_final.json'
for root, dirs, files in os.walk( abs_path ):
for name in files:
if name == json0:
json_path0 = os.path.abspath(os.path.join(root, name))
json_path1 = json_path0.replace('\\','/')
# 2.1 Class A
# query sql command within appropriate functions
Class SqlManager(object):
def __init__(self):
# Set Connection)
self.conn = pymssql.connect(server = sql_host[4:], user = sql_user,
password = sql_password, database = sql_db)
def __del__(self):
def GetRecentProcData(self):
def MonitorAdb(self):
def MonitorBdb(self):
def UpdateResultAdb(self):
def UpdateResultBdb(self):
# 2.2 Class B
Class PreprocDL(object):
def __init__(self, json_path1):
def SetModel(self):
def Normalize(self, data, w, h):
def PosToIndex(self, pos):
def ReadFile(self, identifier, data, additional):
def PreProcess(self, identifier, data, additional):
def ImageSaver(self, identifier, data, additional):
def DoClassify(self, identifier, data, additional):
# 2.3 Function C
def scheduler1():
# Load a function in Class A, which monitor Database
...
# if NewData is not empyty, execute pre-processing and prediction with loaded model
...
# run timer
thread0 = threading.Timer(CHK_TIME, scheduler1)
thread0.start()
# 3. Main funciton
CHK_TIME = 10
if __name__ == "__main__":
# SQL Manager
SqlManager = SqlManager()
# Classifier
MyClf = PreprocDL(json_path1)
# Load trained model paramerters
init_res = MyClf.SetModel()
# SetModel() returns init_res as an indicator of model loading by Catch
if init_res == 0:
try :
scheduler()
except Exception as err:
logging.exception("Error!")
else :
logging.debug("\nDeep Learning Model Loading Fail...!\n" )
# raise Exception
raise RuntimeErrorBuild executable file(.exe) using pyinstaller
Things to remember
- If Tensorflow environment is CPU(GPU) based, trained model should be built upon CPU(GPU)
- Type in required packages when building an executable file
Example of generating one python executable file in command line.
pyinstaller –hidden-import=scipy.integrate –hidden-import=scipy.integrate.quadpack –hidden-import=scipy._lib.messagestream –hidden-import=scipy.integrate._vode -F filename.py
Multi-GPUs
Cases where Multiple GPUs are beneficial
When I encountered to work on multiple projects at the same time, I was supported for using a workstation with 4 GPUs(GeForce 1080Ti, 11G). While delving in to find out the way to efficiently utilize given resources, I figured out that Tensorflow offers some options that enable us to assign multiple GPUs. Below contents describe how to assign GPUs corresponding to your intention, tasks, and models. Specifically, there could be 3 cases. First, multiple jobs(scripts) at different GPUs. Secondly, multiple models at each GPU. Finally, one big model to distributed GPUs.
1. Multi tasks –> Multi GPUs
When you have multiple projects(scripts) and each project requires to use GPU, you can assign each GPU to the corresponding project(script). Without declaration about the list of the visible device, Tensorflow captures all accessible GPUs. To prevent such holdings, assigning Configuration with GPU naming in Session is necessary. Suppose that model is already declared and training with specified epoch will be followed.
# ===== Script A.py ===== #
# Configuration Declaration
a = tf.ConfigProto()
# Assign GPU number to Configuration
a.gpu_options.visible_device_list= "0"
# Finally, allocate Configuration to Session
sess = tf.Session(config=a)
init = tf.global_variables_initializer()
sess.run(init)
# ===== Script B.py ===== #
# Configuration Declaration
b = tf.ConfigProto()
# Assign GPU number to Configuration
b.gpu_options.visible_device_list= "1"
# Finally, allocate Configuration to Session
sess = tf.Session(config=b)
init = tf.global_variables_initializer()
sess.run(init)2. Multi models –> Multi GPUs
If implementing multiple models at each GPUs(e.g. ensemble) is required, each GPU can be allocated to models(graph). Additionally, if each graph shares lots of common parts such as input, output, dropout, and cost, declare graph with “scope” allow us to access each component of graph more efficiently at training parts. In order to show how to access shared component name across the different graph, I only changed the size of the node at the last layer of each graph and named components same. Although runtime might vary depending on the size of batch and data, training graph parallel across GPUs shorten the time about 1 sec for each iteration rather than training each graph sequentially(e.g. MNIST data with 10 batch size is used).
scopes = []
# ==== ==== Grpah 1 Declaration
graph1 = tf.Graph()
with graph1.as_default():
with graph1.name_scope( "g1" ) as scope:
scopes.append( "g1" + "/" )
X = tf.placeholder("float", [None, 28, 28, 1], name = "input")
Y = tf.placeholder("float", [None, 10], name = "label")
w = init_weights([3, 3, 1, 64]) # 3x3x1 conv, 32 outputs
w2 = init_weights([3, 3, 64, 64]) # 3x3x32 conv, 64 outputs
w3 = init_weights([3, 3, 64, 128]) # 3x3x32 conv, 128 outputs
w4 = init_weights([128 * 4 * 4, 2400]) # FC 128 * 4 * 4 inputs, 2400 outputs
w_o = init_weights([2400, 10]) # FC 2400 inputs, 10 outputs (labels)
p_keep_conv = tf.placeholder("float", name = "p_keep_conv")
p_keep_hidden = tf.placeholder("float", name = "p_keep_hidden")
py_x = model(X, w, w2, w3, w4, w_o, p_keep_conv, p_keep_hidden)
cost = tf.reduce_mean(tf.nn.softmax_cross_entropy_with_logits(logits = py_x, labels = Y), name = "cross_entropy" )
train_op = tf.train.RMSPropOptimizer(0.001, 0.9).minimize(cost, name = "train_step" )
predict_op = tf.argmax(py_x, 1, name = "predict_op")
initialize = tf.global_variables_initializer()
# Grpah 2 Declaration
graph2 = tf.Graph()
with graph2.as_default():
with graph2.name_scope( "g2" ) as scope:
scopes.append( "g2" + "/" )
X = tf.placeholder("float", [None, 28, 28, 1], name = "input")
Y = tf.placeholder("float", [None, 10], name = "label")
w = init_weights([3, 3, 1, 64]) # 3x3x1 conv, 32 outputs
w2 = init_weights([3, 3, 64, 64]) # 3x3x32 conv, 64 outputs
w3 = init_weights([3, 3, 64, 128]) # 3x3x32 conv, 128 outputs
w4 = init_weights([128 * 4 * 4, 4800]) # FC 128 * 4 * 4 inputs, 4800 outputs
w_o = init_weights([4800, 10]) # FC 4800 inputs, 10 outputs (labels)
p_keep_conv = tf.placeholder("float", name = "p_keep_conv")
p_keep_hidden = tf.placeholder("float", name = "p_keep_hidden")
py_x = model(X, w, w2, w3, w4, w_o, p_keep_conv, p_keep_hidden)
cost = tf.reduce_mean(tf.nn.softmax_cross_entropy_with_logits(logits = py_x, labels = Y), name = "cross_entropy" )
train_op = tf.train.RMSPropOptimizer(0.001, 0.9).minimize(cost, name = "train_step" )
predict_op = tf.argmax(py_x, 1, name = "predict_op")
initialize2 = tf.global_variables_initializer()
# Session for each graph and GPU
sessions = []
b = tf.ConfigProto()
b.gpu_options.visible_device_list= "1"
sess1 = tf.Session(graph=graph1,config=b)
sess1.run(initialize)
sessions.append(sess1)
c = tf.ConfigProto()
c.gpu_options.visible_device_list= "2"
sess2 = tf.Session(graph=graph2,config=c)
sess2.run(initialize)
sessions.append(sess2)
# Training by matching Sessions to Grpah with GPUs
for i in range(1000):
for j in range(len(sessions)):
training_batch = zip(range(0, len(trX), batch_size), range(batch_size, len(trX)+1, batch_size))
for start, end in training_batch:
ys = sessions[j].run([ sessions[j].graph.get_tensor_by_name( scopes[j] + 'cross_entropy:0' ) ],
feed_dict={sessions[j].graph.get_tensor_by_name( scopes[j] + 'input:0' ): trX[start:end],
sessions[j].graph.get_tensor_by_name( scopes[j] + 'label:0' ): trY[start:end],
sessions[j].graph.get_tensor_by_name( scopes[j] + 'p_keep_conv:0' ): 0.8,
sessions[j].graph.get_tensor_by_name( scopes[j] + 'p_keep_hidden:0' ): 0.75})
test_indices = np.arange(len(teX)) # Get A Test Batch
np.random.shuffle(test_indices)
test_indices = test_indices[0:test_size]
if j == 0 :
print("Running on GPU # 1 ")
else:
print("Running on GPU # 2 ")
print(i, np.mean(np.argmax(teY[test_indices], axis=1) == sessions[j].run(sessions[j].graph.get_tensor_by_name( scopes[j] + 'predict_op:0' ) ,
feed_dict={sessions[j].graph.get_tensor_by_name( scopes[j] + 'input:0' ): teX[test_indices],
sessions[j].graph.get_tensor_by_name( scopes[j] + 'label:0' ): teY[test_indices],
sessions[j].graph.get_tensor_by_name( scopes[j] + 'p_keep_conv:0' ): 1.,
sessions[j].graph.get_tensor_by_name( scopes[j] + 'p_keep_hidden:0' ): 1.}) ) )3. One big model –> Distributed GPUs
Once the size of necessary model exceeds that of GPU memory, the model can be divided into multiple jobs where each of them are executed by different GPUs.
#
graph1 = tf.Graph()
with graph1.as_default():
# GPU Assignments within a Graph
with tf.device( '/gpu:1' ) :
X = tf.placeholder("float", [None, 28, 28, 1], name = "input")
Y = tf.placeholder("float", [None, 10], name = "label")
w = init_weights([3, 3, 1, 64]) # 3x3x1 conv, 32 outputs
w2 = init_weights([3, 3, 64, 64]) # 3x3x32 conv, 64 outputs
w3 = init_weights([3, 3, 64, 128]) # 3x3x32 conv, 128 outputs
with tf.device( '/gpu:2' ) :
w4 = init_weights([128 * 4 * 4, 2400]) # FC 128 * 4 * 4 inputs, 2400 outputs
w_o = init_weights([2400, 10]) # FC 2400 inputs, 10 outputs (labels)
p_keep_conv = tf.placeholder("float", name = "p_keep_conv")
p_keep_hidden = tf.placeholder("float", name = "p_keep_hidden")
py_x = model(X, w, w2, w3, w4, w_o, p_keep_conv, p_keep_hidden)
cost = tf.reduce_mean(tf.nn.softmax_cross_entropy_with_logits(logits = py_x, labels = Y), name = "cross_entropy" )
train_op = tf.train.RMSPropOptimizer(0.001, 0.9).minimize(cost, name = "train_step" )
predict_op = tf.argmax(py_x, 1, name = "predict_op")
# Assign a Graph to session
with tf.Session(graph = graph1) as sess:
tf.global_variables_initializer().run()
for i in range(100):
training_batch = zip(range(0, len(trX), batch_size), range(batch_size, len(trX)+1, batch_size))
for start, end in training_batch:
sess.run(train_op, feed_dict={X: trX[start:end], Y: trY[start:end], p_keep_conv: 0.8, p_keep_hidden: 0.5})
test_indices = np.arange(len(teX)) # Get A Test Batch
np.random.shuffle(test_indices)
test_indices = test_indices[0:test_size]
print(i, np.mean(np.argmax(teY[test_indices], axis=1) ==
sess.run(predict_op, feed_dict={X: teX[test_indices],
Y: teY[test_indices],
p_keep_conv: 1.0,
p_keep_hidden: 1.0})))Wrap up
Hopefully, I wish to improve the performance of carrying out the code by properly applying described methods with multi-GPU. On the other hand, although GPU seems to be powerful for computation-intensive works, it might slow down for the case of running conditional statements relative to CPU. Sp, optimally assigning each task or job to appropriate memory can be another option to boost the performance of work.

